|
Frequently Asked Questions
1.
What is a gene trap?
2.
How do gene trap vectors work?
3.
How can I find out if a mutation in my gene of interest already
exists?
4.
I found my gene! What should I do now?
1. What is a gene trap?
Gene trapping creates random insertional mutations in the genome
that are immediately accessible to molecular characterization. Gene trap
vectors contain a promoterless reporter gene and/or selectable marker
which is activated upon insertion within an endogenous gene, hence,
"gene trapping". The gene trap vector itself provides a DNA tag
for the rapid identification of the disrupted locus. When carried
out in mouse embryonic stem (ES) cells, gene trap insertions can
be transmitted to the germline of mice to examine the function of
the trapped gene in vivo. In contrast to other mutagenesis methods
such as the use of ENU, gene traps generally induce null mutations.
Gene trapping is a rapid and cost-efficient method that is ideally
suited for large-scale mutagenesis of the mammalian genome. Mutations
are relatively inexpensive to create and mutant cell lines are easily
stored and distributed. The IGTC is committed to creating a large
public resource of gene trap insertions that can be used to analyse
gene function in mice. This resource is intended to provide all
investigators equal access to knock-out mice and thereby accelerate
the rate by which new mutant strains of mice can be generated for
research purposes.
2. How do gene trap vectors
work?
Two types of vectors are commonly used, each of which
can be introduced by electroporation or retroviral infection (Figure
1). The "intron trap" includes a splice acceptor sequence immediately
upstream of a promoterless reporter gene that is activated following
insertions in introns of genes. The "exon trap" lacks a splice
acceptor and is designed to activate the reporter following insertions
in exons.
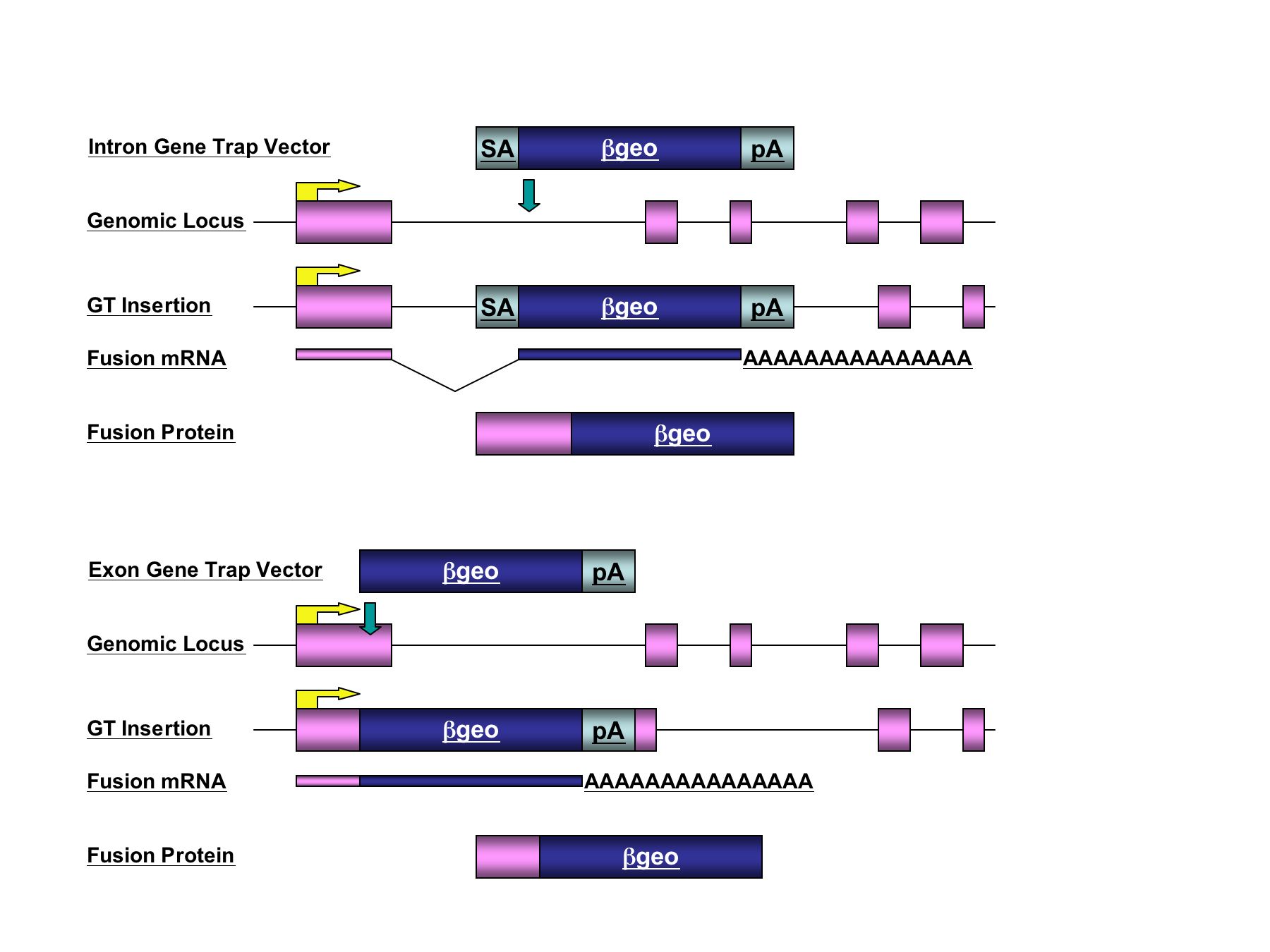
Figure
1. Integration within an endogenous gene places the reporter gene
under the transcriptional control of the "trapped" gene's promoter.
A fusion transcript is generated between upstream exons and the
reporter gene. The polyadenylation signal (pA) within the vector
defines the final exon of the endogenous transcription unit.
Gene trap insertions have three important features:
 |
The vector sequences provide a molecular tag for characterization
of the disrupted gene. Amplification of the fusion transcript
by RACE (Rapid Amplification of cDNA Ends) or amplification of
flanking genomic sequence by inverse PCR is used to generate a
sequence tag for each trapped gene. |
 |
The expression pattern of the trapped gene can be followed by
monitoring reporter gene activity, both in vivo and in vitro (e.g.
by staining for galactosidase activity). Knowing the expression
pattern of the trapped gene is useful for the analysis of the
mutant phenotype. |
|  |
The fusion transcripts generated by gene traps effectively disrupt
gene function by truncating the endogenous mRNA transcript. In
most cases, gene trap insertions create null alleles. Functional
analysis of the gene can proceed following germline transmission
of the gene trap ES cell line.
|
3. How can I find out if a mutation in my gene of interest already
exists?
Most centers have developed their own databases that can
be searched online by keyword or sequence (see Links page). All
gene trap sequence tags are also deposited in the NCBI Genome
Survey Sequences (GSS) database (www.ncbi.nlm.nih.gov/dbGSS/index.html)
and can be searched by BLAST. The IGTC is currently establishing
a unified searchable database with the NCBI. The map position
of each gene trap insertion can be viewed on the Ensembl mouse
genome browser (www.ensembl.org/Mus_musculus)
by selecting the "Gene Trap" feature under 'DAS Source'. Please
check the IGTC home page for updates!
4. I found my gene! What should
I do now?
For most major centres, the sequencing and annotation process
is automated. It is recommended that you download the sequence
tag and confirm for yourself that it has been correctly annotated.
This is most easily done by BLAST at the NCBI or by SSAHA at Ensembl.
To request the cell line(s) from the respective centre, follow
the links on their website. ES cell lines are freely available
to the academic scientific community on a non-collaborative basis
and you will be charged a modest shipping and handling fee to
cover the cost of recovering and preparing cells for shipping.
Finally, it is highly recommended that you confirm that
your gene of interest is disrupted in each ES cell clone before
you attempt to make mice. Each centre should provide information
on the culture of ES cell clones and protocols for confirming
the identity of the trapped gene.
(Text and figures provided by P. Soriano, W. Skarnes and G. Hicks).
| 






